We are proud to host the Bay Area Population Genomics meeting at SF State!
We are proud to host the Bay Area Population Genomics meeting at SF State!

A little while ago we published a new manuscript on fitness costs on the bioRxiv. I’m very excited about this paper, because it is on a new topic for me (fitness costs) and we found some exciting results (for example, I never expected to find that CpG sites were so costly for HIV).
I am also excited about the paper because it is the first paper from my lab at SFSU and it is the first paper that resulted from our collaboration with Adi Stern in Tel Aviv.
The work was done by Marion Hartl (SFSU), Kristof Theys (University of Leuven and SFSU), Alison Feder (Stanford), Maoz Gelbart (University of Tel Aviv), Adi Stern (University of Tel Aviv) and myself.
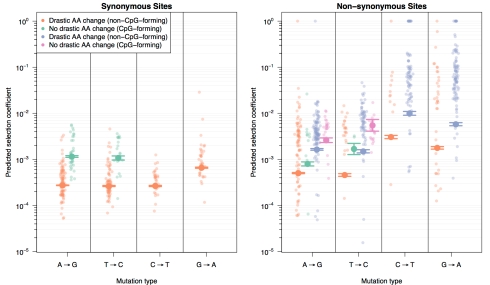
Fig 2 from the manuscript. Selection coefficients for transitions at every nucleotide site in the pol sequence show that CpG-forming mutations are more costly than non-CpG-forming mutations and that mutations that involve a drastic amino acid change are more costly than mutations that do not.
Selection coefficients were estimated using a generalized linear model and sequence data from 160 HIV-infected patients. Shown are predicted selection coefficients for synonymous (left) and non-synonymous (right) mutations that do not involve a drastic amino acid change and either create CpG sites (green) or do not (orange). For non-synonymous mutations, predictions are also shown for mutations that do involve drastic amino acid changes and either create CpG sites (pink) or do not (blue).
Hi Everyone!
Here’s an update about the summer Code Program! Now in week 3, I think everyone is feeling a little bit more comfortable with coding.
In week 1, we worked on a problem set that included vectors, plotting, and rbind/cbind and in week 2, we went through a HIV problem set. This week we’ll be working on some data about ant colonies (Click here to read more about the data: small scale spatial structure of ants of the species Temnothorax longispinosus)!
Also, it has been very nice having socials with Rori’s and Scott’s lab (the other computational labs on campus). We have tried to get together for lunches more and we just had our second social with snacks!
Last Wednesday in the first session ….
We read this press release chosen by one of our students, Christine:
We talked about Figure 1 and the use of DNA sequences versus amino acid sequences and the pros and cons how using either in analysis. Then we broke up into two groups and the students worked on the methods section… one group was to explain what Whole exome sequencing was and the other what RT-PCR was.
You can see the picture attached of the two post-it poster 🙂

After lunch…
DJ did a mini lecture following up on ‘for loops’ from the day before and the ability of writing your own functions in R. Stuff that we have learned and seen in the R book!

Now we are all just working independently! I think the students are learning a lot and are open to asking questions when they need help.
Here is a picture of some of the students in the program!

A lot is happening in the CoDE lab! Here are some updates on what everyone has been up to this spring and what we are planning for the summer!
Marion Hartl
This semester, Marion spent seven weeks in Tel Aviv to work in the lab of our collaborator Adi Stern on improving accuracy of sequencing methods for HIV patient samples. She very much enjoyed being in a country where being a vegetarian is the norm rather than the exception.

Melissa Luk
Melissa is graduating this semester with a Major in Cell and Molecular Biology and a Minor in Chemistry. She is very excited to move to UC Davis next semester to start a Master’s Degree in Forensic Science concentrating on DNA track. She enjoys spending time with her nephew, Gabriel.

Farheen Ghiasuddin
Farheen is graduating this spring with honors as a Physiology Major and a Chemistry Minor. She is planning to take her MCAT soon and then apply for med school in New York. This semester Farheen enjoyed watching The Flash!

Dwayne Evans
Dwayne presented a poster about his graduate research at the CoSE Showcase. He made a lot of progress with his R coding skills this semester. He is preparing to teach a hiphop class soon.

Patricia Kabeja
Patricia is graduating this spring with honors as a Physiology Major and a Chemistry Minor. She is planning to take her MCAT soon and then apply for med school on the East Coast. This summer Patricia is going on a medical mission in Jamaica. Patricia’s favorite character in Game of Thrones is Khaleesi.

Julia Pyko
Julia spent most of this semester interviewing at various medical schools across the country. She received many offers but has not yet selected where she will spend the next 4 years. Her current interests are neurology and OB/GYN. She is getting married this summer and is excited to have all her dogs under one roof again!

Olivia Pham
Olivia will start a Master’s in the fall (in the CoDE lab!) and she got accepted in the prestigious RISE program. Olivia presented a poster at the CoSE Showcase on the work she did as part of the Microbial Genomics class this semester. Olivia plans to learn to play the drums this summer.

Kadie Williams
Kadie presented her research at the CoSE Showcase, and won 4th place in the Graduate Life Science category! Congrats Kadie!! She is the co-founder and president of the Biology Student Council. Currently enjoying her time working on her prospectus and research.
Dasha Fedorova
Dasha is the president of the MEDLIFE chapter at San Francisco State. She has planned a trip, along with her chapter, to go volunteer in Lima, Peru in June. On this trip, she will be setting up health clinics for low-income communities, as well as helping with various developmental projects. She is graduating Fall 2016, and will be taking the MCAT and applying to medical school shortly after.

Sidra Tufon
Sidra is graduating this summer as a Physiology Major and a Chemistry Minor. This is her last year of having the title of a fundraising officer for SFSU’s MEDLIFE chapter. She is planning to take her MCAT next April and then applying to med schools for the next cycle. She will also work as a medical assistant while gaining clinical exposure in medical offices this fall. This summer she plans travel to Cabo and Miami with her friends and family as a graduation treat.

Austin Lim
Austin left the lab early during the spring semester because he found a full time job in a microbiology lab! Congrats Austin!


The Biology and Computer Science departments at SF State are excited to announce a new program: Promoting Inclusivity in Computing (PINC) with the goal of attracting and supporting more Biology students to Computer Science.
A few faculty members from the Biology department have joined forces with the Computer Science department to design a program that lowers the barriers that Biology students experience in learning computer science skills. This program will expose students to basic computing topics such as web design, mobile app development, data structures, and algorithms. All classes in the PINC program are new and especially designed for biologists.
In the PINC program, biology majors will complete 15 units of computer science course work (5 courses spread over 4 semesters) that will allow the students to earn an “Emphasis in Computer Science”.
For more information about PINC, see pincsfsu.com !
The CoDE Lab is very excited to announce a summer coding program that aims to promote more opportunities for undergraduate students to participate in ongoing research!
We have designed a free two-month program that will expose undergraduates not only to research in a lab but also to learn the basics of coding, reading scientific journal articles, and participate in lab meetings. The main goal is for students to learn new skills in the coding language R and be able to apply computational analysis to biological problems.
This program will begin June 7, 2016 and run through August 4, 2016.
Our weekly meetings will consist of coding with learning programs such as Udacity, Code Academy, and worksheets provided by the facilitators, reading journal articles related to drug resistance and other topics of research students may be interested in, and lab meetings. Also, we are hoping to go on field trips to various places where coding in biology is applicable!
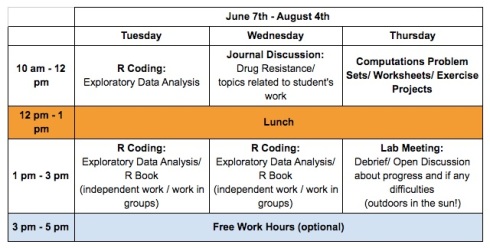
Each week will look roughy like this:
There is four of us organizing the program. We are here to guide and mentor students. Please contact us (phamorrific@gmail.com) if you have any questions. We’d love to hear from you.
Olivia Pham (graduate student in the CoDE lab)

Dwayne Evans (graduate student in the CoDE lab)

Kadie Williams (graduate student in the CoDE lab)

Marion Hartl (Post-doc in the CoDE lab)

This program is for SF State Undergraduates in biology, whom are interested in learning about computational biology, HIV, evolution and/or drug resistance.
Previous experience with programming is not required.
The only requirement for this program is enthusiasm and willingness to work hard. Also, being nice is a plus…! However, we do ask that students who are interested be able to commit at least 6 hours/week during the days that we meet.
If you are not a biology student at SF State but are interested, please contact us and we’ll see what we can do.
This program is free for students, there is no fee to apply!
The program will be held in the CoDE Lab at SF State Hensil Hall 520. This program will begin June 7, 2016 and run through August 4, 2016.
The deadline to apply is May 6th, 2016.
Please fill out the application at: http://goo.gl/forms/7vGrWC4Mtc
Very happy to announce that we have a new paper out and an accompanying video! The paper is about how effective treatments lead to (few) hard selective sweeps and bad treatments lead to soft selective sweeps.
The paper can be found here on the eLife website, but I suggest starting with the video that Alison Feder made.
Title: More effective drugs lead to harder selective sweeps in the evolution of drug resistance in HIV-1.
Authors: Alison F Feder, Soo-Yon Rhee, Susan P Holmes, Robert W Shafer, Dmitri A Petrov, Pleuni S Pennings
DOI: http://dx.doi.org/10.7554/eLife.10670
Abstract: In the early days of HIV treatment, drug resistance occurred rapidly and predictably in all patients, but under modern treatments, resistance arises slowly, if at all. The probability of resistance should be controlled by the rate of generation of resistance mutations. If many adaptive mutations arise simultaneously, then adaptation proceeds by soft selective sweeps in which multiple adaptive mutations spread concomitantly, but if adaptive mutations occur rarely in the population, then a single adaptive mutation should spread alone in a hard selective sweep. Here, we use 6717 HIV-1 consensus sequences from patients treated with first-line therapies between 1989 and 2013 to confirm that the transition from fast to slow evolution of drug resistance was indeed accompanied with the expected transition from soft to hard selective sweeps. This suggests more generally that evolution proceeds via hard sweeps if resistance is unlikely and via soft sweeps if it is likely.
With Ben Wilson, Nandita Garud, Alison Feder and Zoe Assaf, I wrote a review paper about population genetics and drug resistance. It was a lot of fun to write this paper and I feel like I learned a lot during the process.
We wrote about drug resistance in influenza, malaria, TB, MRSA and HIV. It turns out that each of these case studies have something unique to teach us about evolution.
The paper is now out in Molecular Ecology. You can also download it here: 2015Wilson_et_al-Molecular_Ecology.
We made five short movies about the paper. Have a look at the one you are most interested in!
MolEcolNandita from Pleuni Pennings on Vimeo.
BenMolEcol from Pleuni Pennings on Vimeo.
MolEcol from Pleuni Pennings on Vimeo.
MolEcolAlison from Pleuni Pennings on Vimeo.
MolEcolZoe from Pleuni Pennings on Vimeo.
After several brainstorming meetings, we decided on a new name: The CoDE Lab.
CoDE stands for Coding to understand Disease Evolution.
I am very happy with the new name!

The SFSU CoDE Lab in November of 2015. Kristof Theyss, Dasha Fedorova, Sidra Tufon, Patricia Kabeja, Farheen Ghiasuddin, Pleuni Pennings, Melissa Luk, Marton Hartl, Julia Pyko, Olivia Pham, Austin Lim and Dwayne Evans. Not in the picture: Kadie Williams.
Together with several collaborators at Stanford, I published a paper on the BioRxiv on selective sweeps in HIV. We find that when treatments didn’t work well (think AZT in the 1980s), sweeps were very soft, but with better treatments sweeps are getting “harder.” The first author on the paper, Alison Feder, has done most of the work on this paper including making all the figures (and they are cool!).
Alison F Feder, Soo-Yon Rhee, Robert W Shafer, Dmitri A Petrov, Pleuni S Pennings. 2015. More efficacious drugs lead to harder selective sweeps in the evolution of drug resistance in HIV-1. BioRxiv, doi:http://dx.doi.org/10.1101/024109
In the early days of HIV treatment, drug resistance occurred rapidly and predictably in all patients, but under modern treatments, resistance arises slowly, if at all. The probability of resistance should be controlled by the rate of generation of resistant mutations. If many adaptive mutations arise simultaneously, then adaptation proceeds by soft selective sweeps in which multiple adaptive mutations spread concomitantly, but if adaptive mutations occur rarely in the population, then a single adaptive mutation should spread alone in a hard selective sweep. Here we use 6,717 HIV-1 consensus sequences from patients treated with first-line therapies between 1989 and 2013 to confirm that the transition from fast to slow evolution of drug resistance was indeed accompanied with the expected transition from soft to hard selective sweeps. This suggests more generally that evolution proceeds via hard sweeps if resistance is unlikely and via soft sweeps if it is likely.
Figure 3: Drug resistance mutations are correlated with diversity reduction differently in different types of treatments. Treatment efficacy from literature review (% of patients with virologic suppression after 48 weeks) showed positive correspondence with clinical recommendation among RTI regimens (A) and PI+RTI regimens (B). DRM SE lower among the more efficacious and clinically recommended treatments among RTI treatments (C) and RTI+PI treatments (D). Mixed effect model shows significantly different slopes for NNRTI treatments versus NRTI treatments (E) and PI/r treatments versus PI treatments (F). Each line in (EF) represents the fitted decay in diversity with each DRM for a different treatment from the full mixed effects model and p-value labeling indicates the difference between the plotted full model and the null not fitting slopes separately for treatment groups.